Research interests:
Chromosome Structure and Dynamics
Research in my lab is aimed at understanding how (a) nucleoid structure & organization is modulated during cell-cycle; and (b) cohesion regulates homologous recombination processes in E. coli. The long-term goal of my lab is to understand how chromosome dynamics dictates genetic diversity in bacteria.
Chromosome Structure & Organization
High-resolution genome architecture and organization mapping (GANGA): Single locus studies (FROS or FISH) have demonstrated that E. coli chromosome is highly dynamic & fluidic entity. However, spatial mapping of E. coli genome in high-resolution during cell-cycle (G0-S-M) remains an uphill task for cell-biologist. We have been working on “multi-color FISH” (Fluorescence In Situ hybridization) based approach to address this challenge. The approach integrates genetics, biochemical & high-end fluorescence imaging techniques with a MATLAB based image analysis software.
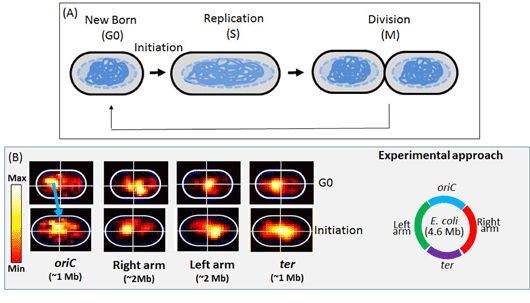
Fig.1. High-Resolution mapping of E. coli nucleoid using GANGA: (a) Schematic representation of E. coli cell-cycle during slow growth. (b) Spatial localization of four different regions (left) of E. coli nucleoid within the cell (schematic, right).
Chromosome cohesion mediated segregation in E. coli
Newly replicated DNA in E. coli is held together by inter-sister linkages before partitioning into daughter nucleoids. Type-II topoisomerase-catalysed sister separation is delayed by the well-characterized cohesin complex in eukaryotes, but cohesion control in E. coli is not currently understood. We propose that SeqA binding results in loose inter-duplex junctions that are resistant to Topo IV cleavage. Epistasis analysis showed that SeqA stabilizes cohesion by antagonizing Topo IV-mediated sister resolution, and possibly also by a direct bridging mechanism. The variable cohesion observed along the E. coli chromosome was due to differential SeqA binding. Reducing cohesion by genetic manipulation of Topo IV or SeqA resulted in dramatically slowed sister locus separation and poor nucleoid partitioning, indicating that cohesion has a prominent role in chromosome segregation.
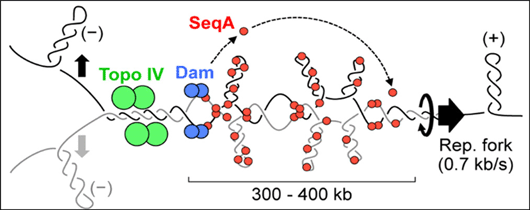
Fig.2. Cohesion mediated segregation of E. coli chromosome: Positive supercoils migrate behind the replisome, entwining newly replicated sister regions. Resolution of precatenanes by Topo IV (green) is delayed by SeqA (red), which binds to hemimethylated DNA tracts behind the fork. Five to ten minutes after fork passage, DNA is remethylated by Dam (blue), releasing SeqA, and allowing Topo IV to resolve inter-sister links.
Chromosome cohesion mediated regulation of Homologous recombination
Homologous recombination (HR) is the major source of antibiotic-resistant gene expansion in pathogenic microbes. HR processes are conserved in all organisms, playing an important role in genomic maintenance during repair of DNA double strand breaks (DSBs) and reactivation of stalled replication fork. However, HR can also induce genomic instability via gene conversion, crossing over and mutation incorporation (under stress), thereby resulting in gene translocations, deletions, amplifications, inversions and loss of heterozygosity. Therefore HR plays a pivotal role in maintaining the equilibrium between genomic integrity and genetic diversity. Although HR is an extensively studied process, it remains unclear how this equilibrium is regulated during DNA repair. Recent data including our own suggested that chromosome cohesion is an evolutionary conserved process and bacteria may also utilize a cohesion dependent mechanism for DSB repair. Therefore, the E. coli provides a highly tractable and mutable model to test the role of cohesion in HR dependent DSB repair.
The focus will be on understanding whether/how (i) cohesion timing along the genome influences the efficiency of DSB repair; (ii) cohesion timing along the genome regulates accumulation of spontaneous and Stress-induced mutation; and (iii) cohesion promotes genomic integrity and dictates the hot-spots for alteration along the genome, in E. coli . This knowledge will be insightful in understanding the mechanism underlying microbial diversity.
|